Persistence
Antibiotic-tolerant persister cells
Bacterial cells use a plethora of mechanisms to evade antibiotic killing. A well-known strategy is antibiotic resistance, in which case the cells have acquired a genomic mutation enabling them to thrive in the presence of the antibiotic. In addition to resistance, every isogenic bacterial population harbours so-called persister cells. These persister cells are phenotypically different from their sensitive kin, and they can withstand even high doses of antibiotics. For a review on how and why persisters can evade antibiotic killing see Wilmaerts, Windels et al. (2019) and Dewachter et al. (2019). The persister phenotype is only temporarily, and upon persister state exit, persisters re-initiate growth and are again susceptible to antibiotics.
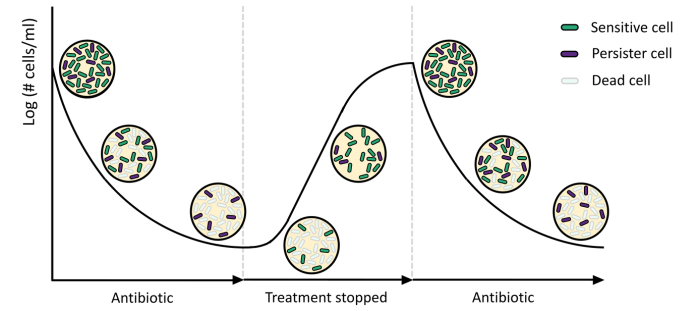
In healthy patients, it is assumed that the immune system can clear the residing persister cells after antibiotic treatment. In immunocompromised patients, or in biofilm-associated infections, persister cells pose a threat as they result in the recolonization of the infection site. Indeed, the recurrence of several chronic infections, such as lung infections in cystic fibrosis patients and tuberculosis is associated with the presence of persister cells (Fauvart, De Groote and Michiels 2011).
In the Michiels lab, persistence of the model bacterium Escherichia coli and of the opportunistic human pathogen Pseudomonas aeruginosa has been studied for many years. Several research lines aim to detect key regulators of persistence, both in E. coli and in P. aeruginosa. Furthermore, these key regulators are subjected to further analysis with the aim of providing a better understanding of the underlying genetic and molecular mechanisms of persister formation (see e.g. Verstraeten, Knapen et al. 2015, Wilmaerts et al. 2018, Wilmaerts et al. 2022, Van den Bergh et al. 2022).
In another project, we perform evolution experiments to select for strains with high persistence levels. On the one hand, this helps us to unravel the genetic basis of persistence. On the other hand, it allows us to determine how persistence evolves in varying antibiotic treatment conditions (see e.g. Van den Bergh et al. 2016). We additionally use experimental evolution to investigate how persistence affects the emergence of genetic resistance (see e.g. Windels, Michiels et al. 2019). The social-evolutionary dimension and ecological impact of persistence are also studied within our group to gather a further fundamental understanding of this phenotype (see e.g. Stepanyan, Wenseleers et al. 2015, Windels et al. 2020, Windels et al. 2021, Verstraete, Van den Bergh et al. 2022).
Meet the team
Philip Reulens, Bram Van den Bergh, Liselot Dewachter, Laure Verstraete, Sang Nguyen, Elen Louwagie, Celien Bollen, Nathan De Boeck, Silke Vercauteren, Giorgio Boccarella, Lieze Agten and Sofie Louwagie are the lab members studying antibiotic-tolerant persister cells.



